Understanding population structure is imperative for sound management and conservation of natural resources. Pacific halibut in Canadian and USA waters are managed as a single, panmictic population on the basis of tagging studies and historical (pre-2010) analyses of genetic population structure that failed to demonstrate significant differentiation in the eastern Pacific Ocean. However, a relatively recent study reported significant genetic population structure suggesting that Pacific halibut residing in the Aleutian Islands may be genetically distinct from other regions examined (Drinan et al. 2016). However, the conclusions from this study required careful consideration becausea relatively small number of genetic markers was used and, more importantly,some of the samples used were collected outside of the spawning season (i.e. winter) and were, therefore, not representative of the local spawning population but rather a mixture of spawning groups on the feeding grounds. Therefore, the IPHC Secretariat has set out to revise current knowledge of population structure of Pacific halibut by using exclusively samples collected during the spawning season and state-of-the-art, high-resolution genomic techniques.
Advances in genomic technology now enable researchers to examine genetic variation at unprecedented resolution. With current techniques the IPHC is now able to move from analyzing a few select genes to entire genomes. Combined with the existing genomic resources that the IPHC has generated, the IPHC is currently using low-coverage whole genome resequencing (lcWGR) (Figure 1) to examine the genetic structure of Pacific halibut in IPHC Convention Waters with unprecedented resolution, adding to our current understanding of Pacific halibut stock structure.
In January and February of 2020, the IPHC conducted genetic sample collections on either side of Amchitka Pass (IPHC Regulatory Area 4B) during the spawning season adding to address the limitations of previous studies. These samples, in combination with samples previously collected during the spawning season in different geographic areas (i.e. Bering Sea, Central Gulf of Alaska and waters off British Columbia) and in different years (1999 to 2007) (Figure 2) will be used to re-evaluate stock structure of Pacific halibut in IPHC Convention waters. The temporal replicates at many of these locations will enable the IPHC to evaluate the stability of genetic structure over time, ensuring confidence in the results. The IPHC has recently produced a high-quality reference genome (Jasonowicz et al. 2022) and has generated genomic sequences from 570 individual Pacific halibut collected from five geographic areas (Figure 1) using lcWGR. Using the lcWGR approach, the IPHC has identified over 10 million single nucleotide polymorphisms (SNPs) that are currently being used to evaluate population structure at the highest resolution possible. With this whole-genome scale dataset we can scan the genome for signals of natural section and/or local adaption and by leveraging IPHC’s existing genome assembly and annotation, we can begin to understand the biological and functional significance of how evolutionary forces shape genetic variation in the Pacific halibut stock.
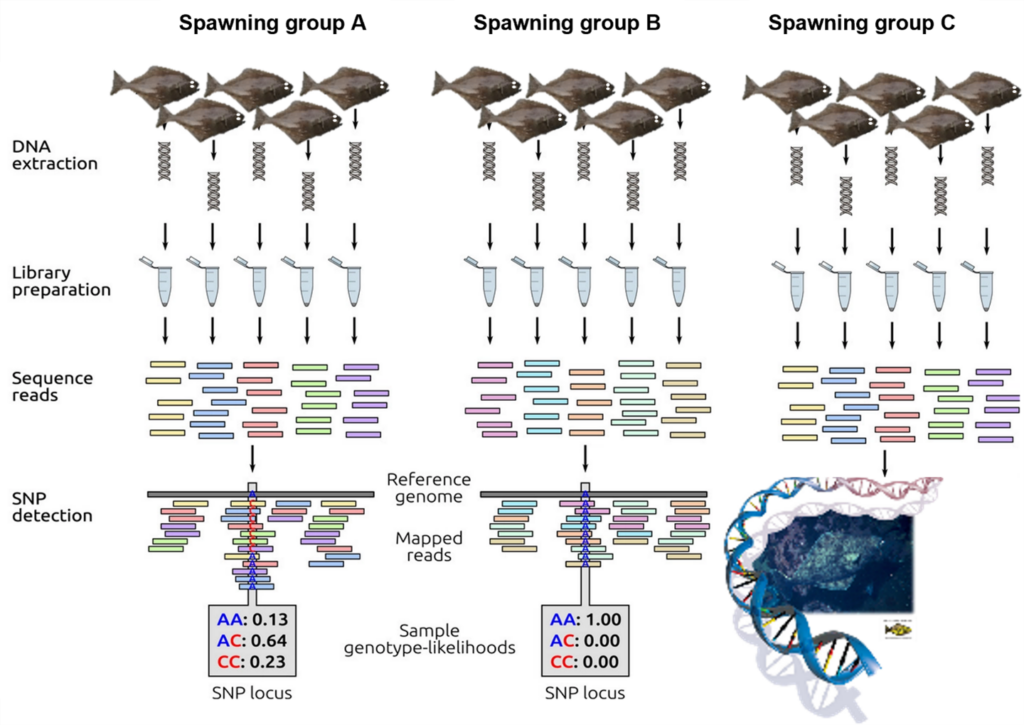
Figure 1. Schematic representation of low-coverage whole-genome resequencing. First DNA is extracted from individual samples and sequencing libraires are prepared using unique barcodes to match DNA sequence reads to individual samples. Sequence reads are mapped to the Pacific halibut reference genome at which point, SNPs can be identified. Figure adapted from Fuentes-Pardo and Ruzzante 2017
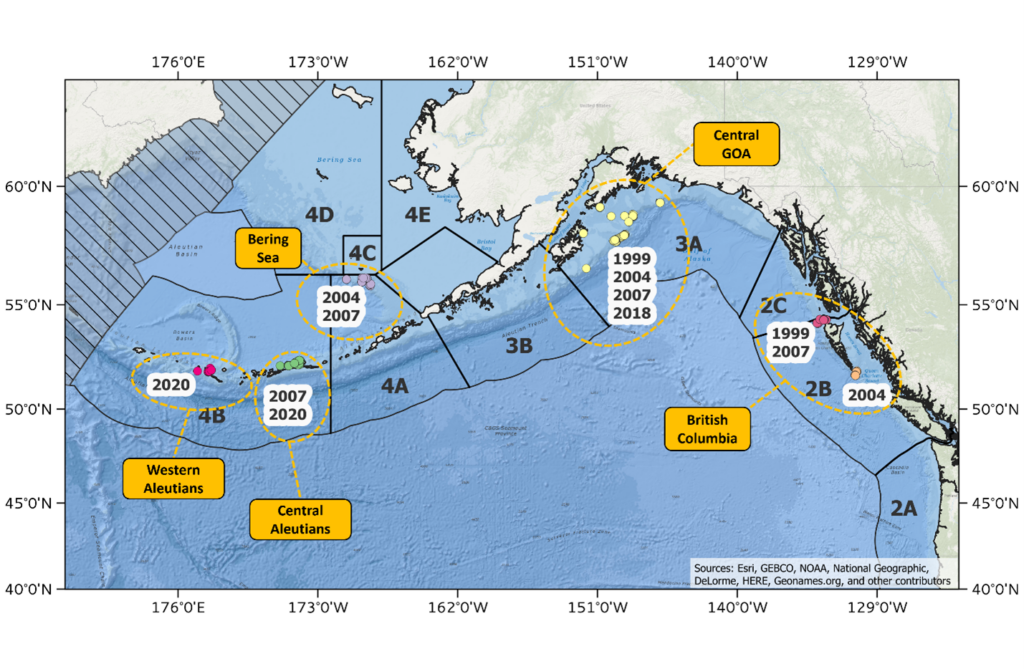
Figure 2. Map of sample collections made during the spawning season used for genomic analysis of population structure in Pacific halibut in the northeast Pacific Ocean.
References:
Drinan, D.P., Galindo, H.M., Loher, T., and Hauser, L. 2016. Subtle genetic population structure in Pacific halibut Hippoglossus stenolepis. Journal of Fish Biology 89(6): 2571–2594. doi:10.1111/jfb.13148.
Fuentes-Pardo, A.P., and Ruzzante, D.E. 2017. Whole-genome sequencing approaches for conservation biology: Advantages, limitations and practical recommendations. Molecular Ecology 26(20): 5369–5406. doi:10.1111/mec.14264.
Jasonowicz, A.J., Simeon, A., Zahm, M., Cabau, C., Klopp, C., Roques, C., Iampietro, C., Lluch, J., Donnadieu, C., Parrinello, H., Drinan, D.P., Hauser, L., Guiguen, Y., and Planas, J.V. 2022. Generation of a chromosome‐level genome assembly for Pacific halibut ( Hippoglossus stenolepis ) and characterization of its sex‐determining genomic region. Molecular Ecology Resources(December 2021): 1–16. doi:10.1111/1755-0998.13641.

